In questo lungo post vi presento il progetto che ho sviluppato per il Data Engineering Nanodegree (DEND) di Udacity. Cosa sviluppare era libera scelta dello sviluppatore posto che alcuni criteri fossero soddisfatti, per esempio lavorare con un database di almeno 3 milioni di records.
Questa è il primo notebook del progetto, nel secondo ci sono esempi di queries che possono essere eseguite sul data lake.
Data lake with Apache Spark¶
Data Engineering Capstone Project¶
Project Summary¶
The Organization for Tourism Development (OTD) want to analyze migration flux in USA, in order to find insights to significantly and sustainably develop the tourism in USA.
To support their core idea they have identified a set of analysis/queries they want to run on the raw data available.
The project deals with building a data pipeline, to go from raw data to the data insights on the migration flux.
The raw data are gathered from different sources, saved in files and made available for download.
The project shows the execution and decisional flow, specifically:
- Chapter 1: Scope the Project and Gather Data. In Step 1 we give more details on the scope of the project.
- Describe the data and how they have been obtained
- Answer the question “how to achieve the target?”
- What infrastructure (storage, computation, communication) has been used and why
- Chapter 2: Explore and Assess the Data. A first look at the data.
- Explore the data
- Check the data for issues, for example null, NaN, or other inconsistencies
- Chapter 3: Define the Data Model. How to organize the data to satisfy the analytical needs.
- Why this data model has been chosen
- How it is implemented
- Chapter 4: Run ELT to Model the Data.
- Load the data from S3 into the SQL database, if any
- Perform quality checks on the database
- Perform example queries
- Chapter 5: Complete Project Write Up
- Documentation of the project
- Possible scenario extensions
# Do all imports and installs here
import pandas as pd
import re
import os
import matplotlib.pyplot as plt
from pyspark.sql import SparkSession
from pyspark.sql.functions import udf, lit, explode, split, regexp_extract, col, isnan, isnull, desc, when, sum, to_date, desc, regexp_replace, count, to_timestamp
from pyspark.sql.types import IntegerType, TimestampType
import boto3
import configparser
#import custom module
from lib import emr_cluster
#setting visualization options
pd.set_option('display.max_colwidth', -1)
pd.set_option('display.max_columns', None)
# modify visualization of the notebook, for easier view
from IPython.core.display import display, HTML
display(HTML("""<style> p { max-width:90% !important; } h1 {font-size:2rem!important } h2 {font-size:1.6rem!important }
h3 {font-size:1.4rem!important } h4 {font-size:1.3rem!important }h5 {font-size:1.2rem!important }h6 {font-size:1.1rem!important }</style>"""))
Index¶
- 1. Scope of the Project
- 1.1 What data
- 1.2 What tools
- 1.3 The I94 immigration data
- 1.3.1 What is an I94?
- 1.3.2 The I94 dataset
- 1.3.3 The SAS date format
- 1.3.4 Loading I94 SAS data
- 1.4 World Temperature Data
- 1.5 Airport Code Table
- 1.6 U.S. City Demographic Data
- 2. Data Exploration
- 2.1 The I94 dataset
- 2.2 I94 SAS data load
- 2.3 Explore I94 data
- 2.4 Cleaning the I94 dataset
- 2.5 Store I94 data as parquet
- 2.6 Airport codes dataset: load, clean, save
- 3. The Data Model
- 3.1 Mapping Out Data Pipelines
- 4. Run Pipelines to Model the Data
- 4.1 Provision the AWS S3 infrastructure
- 4.2 Transfer raw data to S3 bucket
- 4.3 EMR cluster on EC2
- 4.3.1 Provision the EMR cluster
- 4.3.2 Coded fields: I94CIT and I94RES
- 4.3.3 Coded field: I94PORT
- 4.3.4 Data cleaning
- 4.3.5 Save clean data (parquet/json) to S3
- 4.3.6 Loading, cleaning and saving airport codes
- 4.4 Querying data on-the-fly
- 4.5 Querying data using the SQL querying style
- 4.6 Data Quality Checks
- 5. Write up
- Lesson learned
- References
1. Scope of the Project¶
The OTD want to run pre-defined queries on the data, with periodical timing.
They also want to maintain the flexibility to run different queries on the data, using BI tools connected to an SQL-like database.
The core data is the dataset provided by US governative agencies filing request of access in the USA (I94 module).
They also have other lower value data available, that are not part of the core analysis, whose use is unclear, therefore are stored in the data lake for a possible future use.
1.1 What data¶
Following datasets are used in the project:
- I94 immigration data for year 2016. Used for the main analysis
- World Temperature Data
- Airport Code Table
- U.S. City Demographic Data
1.2 What tools¶
Because of the nature of the data and the analysis that must be performed, not time-critical analysis, monthly or weekly batch, the choice fell on a cheaper S3-based data lake with on-demand on-the-fly analytical capability: EMR cluster with Apache Spark, and optionally Apache Airflow for scheduled execution (not implemented here).
The architecture shown below has been implemented.
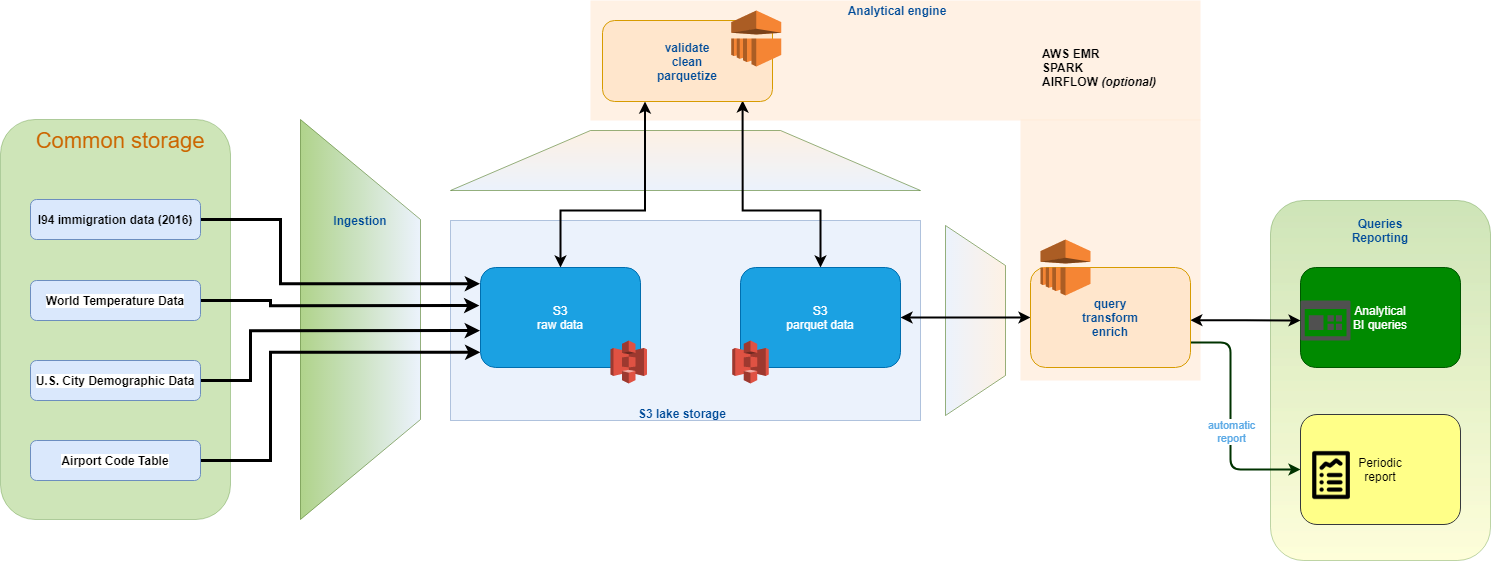
- Starting from a common storage solution (currently Udacity workspace) where both the OTD and its partners have access, the data is then ingested into an S3 bucket, in raw format
- To ease future operations, the data is immediately processed, validated and cleansed using a Spark cluster and stored into S3 in parquet format. Raw and parquet data formats coesist in the data lake.
- By default, the project doesn”t use costly Redshift cluster, but data are queried in-place on the S3 parquet data.
- The EMR cluster serves the analytical needs of the project. SQL based queries are performed using Spark SQL directly on the S3 parquet data
- A Spark job can be triggered monthly, using the Parquet data. The data is aggregated to gain insights on the evolution of the migration flows
1.3 The I94 immigration data¶
The data are provided by the US National Tourism and Trade Office. It is a collection of all I94 that have been filed in 2016.
1.3.1 What is an I94?¶
To give some context is useful to explain what an I94 file is.
From the government website:
“The I-94 is the Arrival/Departure Record, in either paper or electronic format, issued by a Customs and Border Protection (CBP) Officer to foreign visitors entering the United States.”
1.3.2 The I94 dataset¶
Each record contains these fields:
- CICID, unique numer of the file
- I94YR, 4 digit year of the application
- I94MON, Numeric month of the application
- I94CIT, city where the applicant is living
- I94RES, state where the applicant is living
- I94PORT, location (port) where the application is issued
- ARRDATE, arrival date in USA in SAS date format
- I94MODE, how did the applicant arrived in the USA
- I94ADDR, US state where the port is
- DEPDATE is the Departure Date from the USA
- I94BIR, age of applicant in years
- I94VISA, what kind of VISA
- COUNT, used for summary statistics, always 1
- DTADFILE, date added to I-94 Files
- VISAPOST, department of State where where Visa was issued
- OCCUP, occupation that will be performed in U.S.
- ENTDEPA, arrival Flag
- ENTDEPD, departure Flag
- ENTDEPU, update Flag
- MATFLAG, match flag
- BIRYEAR, 4 digit year of birth
- DTADDTO, date to which admitted to U.S. (allowed to stay until)
- GENDER, non-immigrant sex
- INSNUM, INS number
- AIRLINE, airline used to arrive in USA
- ADMNUM, admission Number
- FLTNO, flight number of Airline used to arrive in USA
- VISATYPE, class of admission legally admitting the non-immigrant to temporarily stay in USA
More details in the file I94_SAS_Labels_Descriptions.SAS
1.3.3 The SAS date format¶
Represent any date D0 as the number of days between D0 and the 1th January 1960
1.3.4 Loading I94 SAS data¶
The package saurfang:spark-sas7bdat:2.0.0-s_2.11 and the dependency parso-2.0.8 are needed to read SAS data format.
To load them use the config option spark.jars and give the URL of the repositories, as Spark itself wasn’t able to resolve the dependencies.
spark = SparkSession.builder\
.config("spark.jars","https://repo1.maven.org/maven2/com/epam/parso/2.0.8/parso-2.0.8.jar,https://repos.spark-packages.org/saurfang/spark-sas7bdat/2.0.0-s_2.11/spark-sas7bdat-2.0.0-s_2.11.jar")\
.config("spark.jars.packages","org.apache.hadoop:hadoop-aws:2.7.0")\
.enableHiveSupport()\
.getOrCreate()
1.4 World temperature data¶
The dataset is from Kaggle. It can be found here.
The dataset contains temperature data:
- Global Land and Ocean-and-Land Temperatures (GlobalTemperatures.csv)
- Global Average Land Temperature by Country (GlobalLandTemperaturesByCountry.csv)
- Global Average Land Temperature by State (GlobalLandTemperaturesByState.csv)
- Global Land Temperatures By Major City (GlobalLandTemperaturesByMajorCity.csv)
- Global Land Temperatures By City (GlobalLandTemperaturesByCity.csv)
Below is a snapshop of the global land temperatures by city file.
1.5 Airport codes data¶
This is a table of airport codes, and information on the corresponding cities, like gps coordinates, elevation, country, etc. It comes from Datahub website.
Below is a snapshot of the data.
1.6 U.S. City Demographic Data¶
The dataset comes from OpenSoft. It can be found here.
It contains demographic info on US cities. The info are organized like in the picture below.
2. Data Exploration¶
In this chapter we proceed identifying data quality issues, like missing values, duplicate data, etc.
The purpose is to identify the flow in the data pipeline to programmatically correct data issues.
In this step we work on local data.
2.1 The I94 dataset¶
- How many files are in the I94 dataset?
I94_DATASET_PATH = '../../data/18-83510-I94-Data-2016/'
filelist = os.listdir(I94_DATASET_PATH)
print("The dataset contains {} files".format(len(filelist)))
The dataset contains 12 files
- What is the size of the files?
for file in filelist:
size = os.path.getsize('{}/{}'.format(I94_DATASET_PATH, file))
print('{} - dim(bytes): {}'.format(file, size))
i94_apr16_sub.sas7bdat - dim(bytes): 471990272 i94_sep16_sub.sas7bdat - dim(bytes): 569180160 i94_nov16_sub.sas7bdat - dim(bytes): 444334080 i94_mar16_sub.sas7bdat - dim(bytes): 481296384 i94_jun16_sub.sas7bdat - dim(bytes): 716570624 i94_aug16_sub.sas7bdat - dim(bytes): 625541120 i94_may16_sub.sas7bdat - dim(bytes): 525008896 i94_jan16_sub.sas7bdat - dim(bytes): 434176000 i94_oct16_sub.sas7bdat - dim(bytes): 556269568 i94_jul16_sub.sas7bdat - dim(bytes): 650117120 i94_feb16_sub.sas7bdat - dim(bytes): 391905280 i94_dec16_sub.sas7bdat - dim(bytes): 523304960
2.2 I94 SAS data load¶
To read SAS data format I need to specify the com.github.saurfang.sas.spark format.
I94_TEST_FILE = '../../data/18-83510-I94-Data-2016/i94_apr16_sub.sas7bdat'
df_I94 = spark.read.format('com.github.saurfang.sas.spark').load(I94_TEST_FILE).persist()
df_I94.limit(5).toPandas().head()
| cicid | i94yr | i94mon | i94cit | i94res | i94port | arrdate | i94mode | i94addr | depdate | i94bir | i94visa | count | dtadfile | visapost | occup | entdepa | entdepd | entdepu | matflag | biryear | dtaddto | gender | insnum | airline | admnum | fltno | visatype | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 6.0 | 2016.0 | 4.0 | 692.0 | 692.0 | XXX | 20573.0 | NaN | None | NaN | 37.0 | 2.0 | 1.0 | None | None | None | T | None | U | None | 1979.0 | 10282016 | None | None | None | 1.897628e+09 | None | B2 |
| 1 | 7.0 | 2016.0 | 4.0 | 254.0 | 276.0 | ATL | 20551.0 | 1.0 | AL | NaN | 25.0 | 3.0 | 1.0 | 20130811 | SEO | None | G | None | Y | None | 1991.0 | D/S | M | None | None | 3.736796e+09 | 00296 | F1 |
| 2 | 15.0 | 2016.0 | 4.0 | 101.0 | 101.0 | WAS | 20545.0 | 1.0 | MI | 20691.0 | 55.0 | 2.0 | 1.0 | 20160401 | None | None | T | O | None | M | 1961.0 | 09302016 | M | None | OS | 6.666432e+08 | 93 | B2 |
| 3 | 16.0 | 2016.0 | 4.0 | 101.0 | 101.0 | NYC | 20545.0 | 1.0 | MA | 20567.0 | 28.0 | 2.0 | 1.0 | 20160401 | None | None | O | O | None | M | 1988.0 | 09302016 | None | None | AA | 9.246846e+10 | 00199 | B2 |
| 4 | 17.0 | 2016.0 | 4.0 | 101.0 | 101.0 | NYC | 20545.0 | 1.0 | MA | 20567.0 | 4.0 | 2.0 | 1.0 | 20160401 | None | None | O | O | None | M | 2012.0 | 09302016 | None | None | AA | 9.246846e+10 | 00199 | B2 |
- Let’s see the schema Spark applied on reading the file
df_I94.printSchema()
root |-- cicid: double (nullable = true) |-- i94yr: double (nullable = true) |-- i94mon: double (nullable = true) |-- i94cit: double (nullable = true) |-- i94res: double (nullable = true) |-- i94port: string (nullable = true) |-- arrdate: double (nullable = true) |-- i94mode: double (nullable = true) |-- i94addr: string (nullable = true) |-- depdate: double (nullable = true) |-- i94bir: double (nullable = true) |-- i94visa: double (nullable = true) |-- count: double (nullable = true) |-- dtadfile: string (nullable = true) |-- visapost: string (nullable = true) |-- occup: string (nullable = true) |-- entdepa: string (nullable = true) |-- entdepd: string (nullable = true) |-- entdepu: string (nullable = true) |-- matflag: string (nullable = true) |-- biryear: double (nullable = true) |-- dtaddto: string (nullable = true) |-- gender: string (nullable = true) |-- insnum: string (nullable = true) |-- airline: string (nullable = true) |-- admnum: double (nullable = true) |-- fltno: string (nullable = true) |-- visatype: string (nullable = true)
The most columns are categorical data, this means the information is coded, for example I94CIT=101, 101 is the country code for Albania.
Other columns represent integer data.
It appears clear that there is no need to have data that are defined as double => let’s change those fields to integer
toInt = udf(lambda x: int(x) if x!=None else x, IntegerType())
for colname, coltype in df_I94.dtypes:
if coltype == 'double':
df_I94 = df_I94.withColumn(colname, toInt(colname))
Verifying the schema is correct.
df_I94.printSchema()
root |-- cicid: integer (nullable = true) |-- i94yr: integer (nullable = true) |-- i94mon: integer (nullable = true) |-- i94cit: integer (nullable = true) |-- i94res: integer (nullable = true) |-- i94port: string (nullable = true) |-- arrdate: integer (nullable = true) |-- i94mode: integer (nullable = true) |-- i94addr: string (nullable = true) |-- depdate: integer (nullable = true) |-- i94bir: integer (nullable = true) |-- i94visa: integer (nullable = true) |-- count: integer (nullable = true) |-- dtadfile: string (nullable = true) |-- visapost: string (nullable = true) |-- occup: string (nullable = true) |-- entdepa: string (nullable = true) |-- entdepd: string (nullable = true) |-- entdepu: string (nullable = true) |-- matflag: string (nullable = true) |-- biryear: integer (nullable = true) |-- dtaddto: string (nullable = true) |-- gender: string (nullable = true) |-- insnum: string (nullable = true) |-- airline: string (nullable = true) |-- admnum: integer (nullable = true) |-- fltno: string (nullable = true) |-- visatype: string (nullable = true)
- convert string columns dtadfile and dtaddto to date type
These fields come in a simple string format. To be able to run time-based queries they are converted to date type
df_I94 = df_I94.withColumn('dtadfile', to_date(col('dtadfile'), format='yyyyMMdd'))\
.withColumn('dtadddto', to_date(col('dtaddto'), format='MMddyyyy'))
- convert columns arrdate and depdate from SAS-date format to a timestamp type.
A date in SAS format is simply the number of days between the chosen date and the reference date (01-01-1960)
@udf(TimestampType())
def to_timestamp_udf(x):
try:
return pd.to_timedelta(x, unit='D') + pd.Timestamp('1960-1-1')
except:
return pd.Timestamp('1900-1-1')
df_I94 = df_I94.withColumn('arrdate', to_date(to_timestamp_udf(col('arrdate'))))\
.withColumn('depdate', to_date(to_timestamp_udf(col('depdate'))))
df_I94.limit(5).toPandas().head(5)
| cicid | i94yr | i94mon | i94cit | i94res | i94port | arrdate | i94mode | i94addr | depdate | i94bir | i94visa | count | dtadfile | visapost | occup | entdepa | entdepd | entdepu | matflag | biryear | dtaddto | gender | insnum | airline | admnum | fltno | visatype | dtadddto | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 2016 | 4 | 692 | 692 | XXX | 2016-04-29 | NaN | None | 1900-01-01 | 37 | 2 | 1 | None | None | None | T | None | U | None | 1979 | 10282016 | None | None | None | 1897628485 | None | B2 | 2016-10-28 |
| 1 | 7 | 2016 | 4 | 254 | 276 | ATL | 2016-04-07 | 1.0 | AL | 1900-01-01 | 25 | 3 | 1 | 2013-08-11 | SEO | None | G | None | Y | None | 1991 | D/S | M | None | None | -558170966 | 00296 | F1 | None |
| 2 | 15 | 2016 | 4 | 101 | 101 | WAS | 2016-04-01 | 1.0 | MI | 2016-08-25 | 55 | 2 | 1 | 2016-04-01 | None | None | T | O | None | M | 1961 | 09302016 | M | None | OS | 666643185 | 93 | B2 | 2016-09-30 |
| 3 | 16 | 2016 | 4 | 101 | 101 | NYC | 2016-04-01 | 1.0 | MA | 2016-04-23 | 28 | 2 | 1 | 2016-04-01 | None | None | O | O | None | M | 1988 | 09302016 | None | None | AA | -2020819182 | 00199 | B2 | 2016-09-30 |
| 4 | 17 | 2016 | 4 | 101 | 101 | NYC | 2016-04-01 | 1.0 | MA | 2016-04-23 | 4 | 2 | 1 | 2016-04-01 | None | None | O | O | None | M | 2012 | 09302016 | None | None | AA | -2020817382 | 00199 | B2 | 2016-09-30 |
- print final schema
df_I94.printSchema()
root |-- cicid: integer (nullable = true) |-- i94yr: integer (nullable = true) |-- i94mon: integer (nullable = true) |-- i94cit: integer (nullable = true) |-- i94res: integer (nullable = true) |-- i94port: string (nullable = true) |-- arrdate: date (nullable = true) |-- i94mode: integer (nullable = true) |-- i94addr: string (nullable = true) |-- depdate: date (nullable = true) |-- i94bir: integer (nullable = true) |-- i94visa: integer (nullable = true) |-- count: integer (nullable = true) |-- dtadfile: date (nullable = true) |-- visapost: string (nullable = true) |-- occup: string (nullable = true) |-- entdepa: string (nullable = true) |-- entdepd: string (nullable = true) |-- entdepu: string (nullable = true) |-- matflag: string (nullable = true) |-- biryear: integer (nullable = true) |-- dtaddto: string (nullable = true) |-- gender: string (nullable = true) |-- insnum: string (nullable = true) |-- airline: string (nullable = true) |-- admnum: integer (nullable = true) |-- fltno: string (nullable = true) |-- visatype: string (nullable = true) |-- dtadddto: date (nullable = true)
2.3 Explore I94 data¶
- How many rows does the I94 database has?
df_I94.count()
3096313
- Let’s see the gender distribution of the applicants
df_I94.select("gender").groupBy("gender").count().show()
+------+-------+ |gender| count| +------+-------+ | F|1302743| | null| 414269| | M|1377224| | U| 467| | X| 1610| +------+-------+
- Where are the I94 applicants coming from?
I want to know the 10 most represented nations
df_I94_count_visitors = df_I94.select("i94res").groupby("i94res").count().sort(col("count").desc()).persist()
df_I94_count_visitors.show(10)
+------+------+ |i94res| count| +------+------+ | 135|368421| | 209|249167| | 245|185609| | 111|185339| | 582|179603| | 112|156613| | 276|136312| | 689|134907| | 438|112407| | 213|107193| +------+------+ only showing top 10 rows
The i94res code 135, where the highest number of visitors come from, corresponds to the the United Kingdom, as can be read in the accompanying file I94_SAS_Labels_Descriptions.SAS
- What port registered the highest number of arrivals?
df_I94_total_arrivals_per_port = df_I94.groupBy('i94port').count().sort(col('count').desc()).persist()
df_I94_total_arrivals_per_port.show(10)
+-------+------+ |i94port| count| +-------+------+ | NYC|485916| | MIA|343941| | LOS|310163| | SFR|152586| | ORL|149195| | HHW|142720| | NEW|136122| | CHI|130564| | HOU|101481| | FTL| 95977| +-------+------+ only showing top 10 rows
New York City port registered the highest number of arrivals.
2.4 Cleaning the I94 dataset¶
These are the steps to perform on the I94 database:
- Identify null and NaN values. Remove duplicates (quality check).
- Find errors in the records (quality check) for example dates not in year 2016
- Counting how many NaN in each column, excluding the date type columns dtadfile, dtadddto, arrdate, depdate because the isnan function works only on numerical types
df_I94.select([count(when(isnan(colname), 1)).alias(colname) for colname in df_I94.drop('dtadfile','dtadddto','arrdate','depdate').columns]).toPandas().head()
| cicid | i94yr | i94mon | i94cit | i94res | i94port | i94mode | i94addr | i94bir | i94visa | count | visapost | occup | entdepa | entdepd | entdepu | matflag | biryear | dtaddto | gender | insnum | airline | admnum | fltno | visatype | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
- How many rows of the I94 database have null value?
df_I94.na.fill(False).count()
3096313
The number of nulls equal the number of rows. It means there is at least one null on each row of the dataframe.
- Now we can count how many null there are in each row
%%time
df_I94.select([count(when(col(colname).isNull(),1)).alias('sum_'+colname) for colname in df_I94.columns]).toPandas().head()
CPU times: user 101 ms, sys: 24.4 ms, total: 126 ms Wall time: 8min 13s
| sum_cicid | sum_i94yr | sum_i94mon | sum_i94cit | sum_i94res | sum_i94port | sum_arrdate | sum_i94mode | sum_i94addr | sum_depdate | sum_i94bir | sum_i94visa | sum_count | sum_dtadfile | sum_visapost | sum_occup | sum_entdepa | sum_entdepd | sum_entdepu | sum_matflag | sum_biryear | sum_dtaddto | sum_gender | sum_insnum | sum_airline | sum_admnum | sum_fltno | sum_visatype | sum_dtadddto | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 239 | 152592 | 0 | 802 | 0 | 0 | 1 | 1881250 | 3088187 | 238 | 138429 | 3095921 | 138429 | 802 | 477 | 414269 | 2982605 | 83627 | 0 | 19549 | 0 | 45824 |
There are many nulls in many columns.
The question is, if there is a need to correct/fill those nulls.
Looking at the data, it seems like some field have been left empty for lack of information.
Because these are categorical data there is no use, at this step, in assigning arbitrary values to the nulls.
The nulls are not going to be filled apriori, but only if a specific need comes up.
- Are there duplicated rows?
count_before = df_I94.count()
count_before
3096313
Dropping duplicate row
df_I94 = df_I94.drop_duplicates()
Cheching if the number changed
count_after = df_I94.count()
count_after
3096313
No row has been dropped => no duplicated row
- Verify that all rows have i94yr column equal 2016
This gives confidence on the consistence of the data
df_I94.where(col('i94yr')==2016).count() == df_I94.count()
True
2.5 Store I94 data as parquet¶
I94 data are stored in parquet format in an S3 bucket, they are partinioned using the fields: year, month
S3_bucket_I94 = 'data/S3bucket_temp/I94_data'
df_I94.write.format('parquet').mode('overwrite').partitionBy('i94yr','i94mon').save(S3_bucket_I94)
2.6 The Airport codes dataset¶
df_airport_codes = spark.read.csv('data/csv/airport-codes_csv.csv', header=True, inferSchema=True)
df_airport_codes.printSchema()
root |-- ident: string (nullable = true) |-- type: string (nullable = true) |-- name: string (nullable = true) |-- elevation_ft: integer (nullable = true) |-- continent: string (nullable = true) |-- iso_country: string (nullable = true) |-- iso_region: string (nullable = true) |-- municipality: string (nullable = true) |-- gps_code: string (nullable = true) |-- iata_code: string (nullable = true) |-- local_code: string (nullable = true) |-- coordinates: string (nullable = true)
A snippet of the data
df_airport_codes.toPandas().head(5)
| ident | type | name | elevation_ft | continent | iso_country | iso_region | municipality | gps_code | iata_code | local_code | coordinates | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 00A | heliport | Total Rf Heliport | 11.0 | NA | US | US-PA | Bensalem | 00A | None | 00A | -74.93360137939453, 40.07080078125 |
| 1 | 00AA | small_airport | Aero B Ranch Airport | 3435.0 | NA | US | US-KS | Leoti | 00AA | None | 00AA | -101.473911, 38.704022 |
| 2 | 00AK | small_airport | Lowell Field | 450.0 | NA | US | US-AK | Anchor Point | 00AK | None | 00AK | -151.695999146, 59.94919968 |
| 3 | 00AL | small_airport | Epps Airpark | 820.0 | NA | US | US-AL | Harvest | 00AL | None | 00AL | -86.77030181884766, 34.86479949951172 |
| 4 | 00AR | closed | Newport Hospital & Clinic Heliport | 237.0 | NA | US | US-AR | Newport | None | None | None | -91.254898, 35.6087 |
How many records?
df_airport_codes.count()
55075
There are no duplicates
df_airport_codes.drop_duplicates().count() == df_airport_codes.count()
True
We discover there are some null fields:
df_airport_codes.dropna().count() == df_airport_codes.count()
False
The nulls are in these colomuns:
df_airport_codes.select([count(when(col(colname).isNull(),1)).alias('sum_'+colname) for colname in df_airport_codes.columns]).toPandas().head()
| sum_ident | sum_type | sum_name | sum_elevation_ft | sum_continent | sum_iso_country | sum_iso_region | sum_municipality | sum_gps_code | sum_iata_code | sum_local_code | sum_coordinates | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 7006 | 0 | 0 | 0 | 5676 | 14045 | 45886 | 26389 | 0 |
No action taken to fill the nulls
Finally, let’s save the data in parquet format in our temporary folder mimicking the S3 bucket.
S3_airport_codes = "data/S3bucket_temp/airport_codes"
df_airport_codes.write.parquet(S3_airport_codes, mode="overwrite")
3. The Data Model¶
The core of the architecture is a data lake, with S3 storage and EMR processing.
The data are stored into S3 in raw and parquet format.
Apache Spark is the tool elected for analytical tasks, therefore all data are loaded into Spark dataframe using a schema-on-read approach.
For SQL queries style on the data, Spark temporary views are generated.
3.1 Mapping Out Data Pipelines¶
- Provision the AWS S3 infrastructure
- Transfer data from the common storage to the S3 lake storage
- Provision an EMR cluster. It runs 2 steps then autoterminate, these are the 2 steps:
3.1 Run a spark job to extract codes from file I94_SAS_Labels_Descriptions.SAS and save to S3
3.2 Data cleaning. Find nan, null, duplicate. Save the clean data to parquet files - Generate reports using Spark query on S3 parquet data
- On-the-fly queries with Spark SQL

config = configparser.ConfigParser()
config.read_file(open('config.cfg'))
KEY = config['AWS']['AWS_ACCESS_KEY_ID']
SECRET = config['AWS']['AWS_SECRET_ACCESS_KEY']
S3_BUCKET = config['S3']['S3_bucket']
Create the bucket if it’s not existing
s3 = boto3.resource('s3',
region_name="us-west-2",
aws_access_key_id = KEY,
aws_secret_access_key = SECRET)
try:
s3.create_bucket(Bucket=S3_BUCKET, CreateBucketConfiguration={
'LocationConstraint': 'us-west-2'})
except Exception as e:
print(e)
An error occurred (BucketAlreadyOwnedByYou) when calling the CreateBucket operation: Your previous request to create the named bucket succeeded and you already own it.
4.2 Transfer raw data to S3 bucket¶
Transfer the data from current shared storage (currently Udacity workspace) to S3 lake storage.
A naive metadata system is implemented. It uses a json file to store basic information on each file added to the S3 bucket:
- file name: file being processed
- added by: user logged as | aws access id
- date added: timestamp of date of processing
- modified on: timestamp of modification time
- notes: any additional information
- access granted to (role or policy): admin | anyone | I94 access policy | weather data access policy |
- expire date: 5 years (default)
SHARED_STORAGE = config['S3']['SHARED_STORAGE']
S3_LAKE_RAWDATA = config['S3']['RAW_DATA']
I94_DATA = config['S3']['I94_DATA']
def addMetadataRawFile(json_filepath, origin_filepath, notes = '', access_granted_to = 'anyone', expire_date = '5y'):
"""
This function stores metadata into a specified file
:json_filepath, json file containing the metadata
:origin_filepath, file which metadata are going to be appended to the metadata json file
:notes individual notes for each file
:access_granted_to, possibly values are secret, admin, anyone, policy_name
:expire_date
"""
import pathlib
import json
import re
import datetime
fname = pathlib.Path(origin_filepath)
if os.path.isfile(json_filepath):
with open(json_filepath) as f:
metadata = json.load(f)
else:
metadata = {}
metadata['rawData'] = []
metadata['rawData'].append({
'filename': re.findall('([^\/]*$)', origin_filepath)[0],
'filename_complete': origin_filepath,
'added_by': KEY,
'date_added': datetime.datetime.timestamp(datetime.datetime.now()),
'modified_on': fname.stat().st_mtime,
'notes': notes,
'access_granted_to': access_granted_to,
'expire_date': expire_date})
with open(json_filepath, 'w') as outfile:
json.dump(metadata, outfile)
def fileTransferToS3(origin_filepath, destination_folder, destination_file=None, json_filepath='./data/metadata/rawDataMetadata.json', notes = '', access_granted_to = 'anyone', expire_date = '5y'):
"""
transfer folder or file to S3 bucket
:origin_filepath, a folder or a file. If a folder is given, all the files inside are transferred to the remote destination_folder.
:destination_folder, folder in the S3 bucket, where to copy the files
:destination_file, it is valid only when origin_filepath is a file. If None, the name of the file in the origin_filepath will be used
:metadata_file, file contains the metadata
"""
import re
#if a directory is passed as origin_filepath parameter all files are copied into the destination_folder
if os.path.isdir(origin_filepath):
filelist = os.walk(origin_filepath)
for root, subFolders, files in filelist:
for file in files:
origin_file_path_name = os.path.join(root, file)
bucket_file_path_name = os.path.join(destination_folder, origin_file_path_name.replace(origin_filepath, ''))
print('Transfering file: ', origin_file_path_name, ' ===> ', S3_BUCKET, '/', bucket_file_path_name, '\n')
s3.meta.client.upload_file(origin_file_path_name, S3_BUCKET, bucket_file_path_name)
addMetadataRawFile(json_filepath, origin_file_path_name, notes, access_granted_to, expire_date)
#if a file is passed as origin_filepath
elif os.path.isfile(origin_filepath):
origin_file_path_name = origin_filepath
if destination_file == None:
filename = re.findall('([^\/]*$)', origin_filepath)[0]
bucket_file_path_name = os.path.join(destination_folder, filename)
else:
bucket_file_path_name = os.path.join(destination_folder, destination_file)
print('Transfering file: ', origin_file_path_name, ' ===> ', S3_BUCKET, '/', bucket_file_path_name, '\n')
s3.meta.client.upload_file(origin_file_path_name, S3_BUCKET, bucket_file_path_name)
addMetadataRawFile(json_filepath, origin_filepath, notes, access_granted_to, expire_date)
These dataset are moved to the S3 lake storage:
- I94 immigration data
- airport codes
- US cities demographics
fileTransferToS3(SHARED_STORAGE, os.path.join(S3_LAKE_RAWDATA, I94_DATA),
json_filepath='./data/metadata/rawDataMetadata.json',
access_granted_to='I94_policy',
expire_date = '20y')
Transfering file: ../../data/I94_SAS_Labels_Descriptions.SAS ===> helptheplanet / raw/i94_data/I94_SAS_Labels_Descriptions.SAS Transfering file: ../../data/18-83510-I94-Data-2016/i94_apr16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_apr16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_sep16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_sep16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_nov16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_nov16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_mar16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_mar16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_jun16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_jun16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_aug16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_aug16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_may16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_may16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_jan16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_jan16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_oct16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_oct16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_jul16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_jul16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_feb16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_feb16_sub.sas7bdat Transfering file: ../../data/18-83510-I94-Data-2016/i94_dec16_sub.sas7bdat ===> helptheplanet / raw/i94_data/18-83510-I94-Data-2016/i94_dec16_sub.sas7bdat
fileTransferToS3('./data/csv/airport-codes_csv.csv', os.path.join(S3_LAKE_RAWDATA, 'csv'),
json_filepath='./data/metadata/rawDataMetadata.json',
access_granted_to='anyone',
expire_date = '3y')
Transfering file: ./data/csv/airport-codes_csv.csv ===> helptheplanet / raw/csv/airport-codes_csv.csv
fileTransferToS3('./data/csv/us-cities-demographics.csv', os.path.join(S3_LAKE_RAWDATA, 'csv'),
json_filepath='./data/metadata/rawDataMetadata.json',
access_granted_to='anyone',
expire_date = '20y')
Transfering file: ./data/csv/us-cities-demographics.csv ===> helptheplanet / raw/csv/us-cities-demographics.csv
fileTransferToS3('./data/csv/temperatureData/', os.path.join(S3_LAKE_RAWDATA, 'csv'),
json_filepath='./data/metadata/rawDataMetadata.json',
access_granted_to='anyone',
expire_date = '5y')
Transfering file: ./data/csv/temperatureData/GlobalLandTemperaturesByMajorCity.csv.zip ===> helptheplanet / raw/csv/GlobalLandTemperaturesByMajorCity.csv.zip Transfering file: ./data/csv/temperatureData/GlobalLandTemperaturesByState.csv.zip ===> helptheplanet / raw/csv/GlobalLandTemperaturesByState.csv.zip Transfering file: ./data/csv/temperatureData/GlobalLandTemperaturesByCity.csv.zip ===> helptheplanet / raw/csv/GlobalLandTemperaturesByCity.csv.zip Transfering file: ./data/csv/temperatureData/GlobalLandTemperaturesByCountry.csv.zip ===> helptheplanet / raw/csv/GlobalLandTemperaturesByCountry.csv.zip
4.3 EMR cluster on EC2¶
An EMR cluster on EC2 instances with Apache Spark preinstalled is used to perform the ELT work.
A 3-nodes cluster of m5.xlarge istances is configured by default in the config.cfg file.
If the performance requires it, the cluster can be scaled up to use more nodes and/or bigger instances.
After the cluster has been created, the steps to execute spark cleaning jobs are added to the EMR job flow, the steps are in separate .py files. These steps are added:
- Spark job 1
- extract I94res, i94cit, i94port codes
- save the codes in a json file in S3
- Spark job 2
- load I94 raw data from S3
- change schema
- data cleaning
- save parquet data to S3
The cluster is set to auto-terminate by default after executing all the steps.
4.3.1 Provision the EMR cluster¶
Create the cluster using the code emr_cluster.py [Ref. 3] and emr_cluster_spark_submit.py and and set the steps to execute spark_script_1 and spark_script_2.
These scripts have already been previously uploaded to a dedicated folder in the project’s S3 bucket, and are accessible from the EMR cluster.
emrCluster = emr_cluster.EMRLoader(config)
spark_script_1 = 'spark_4_emr_codes_extraction.py'
spark_script_2 = 'spark_4_emr_I94_processing.py'
emrCluster.custom_steps = [
{
'Name': 'Run spark_script_1',
'ActionOnFailure': 'TERMINATE_CLUSTER',
'HadoopJarStep': {
'Jar': 'command-runner.jar',
'Args': [
'spark-submit',
'--deploy-mode', 'cluster',
'--py-files', 's3://helptheplanet/code/{}'.format(spark_script_1),
's3://helptheplanet/code/{}'.format(spark_script_1),
]
}
},
{
'Name': 'Run spark_script_2',
'ActionOnFailure': 'TERMINATE_CLUSTER',
'HadoopJarStep': {
'Jar': 'command-runner.jar',
'Args': [
'spark-submit',
'--deploy-mode', 'cluster',
'--py-files', 's3://helptheplanet/code/{}'.format(spark_script_2),
's3://helptheplanet/code/{}'.format(spark_script_2),
]
}
}
]
emrCluster.create_cluster()
2021-08-24 21:26:30,293 - lib.emr_cluster - INFO - {'JobFlowId': 'j-Y4TX4JK0GZC5', 'ResponseMetadata': {'RequestId': '40ed075e-9c08-4d30-96f5-bbe6a48c469b', 'HTTPStatusCode': 200, 'HTTPHeaders': {'x-amzn-requestid': '40ed075e-9c08-4d30-96f5-bbe6a48c469b', 'content-type': 'application/x-amz-json-1.1', 'content-length': '116', 'date': 'Tue, 24 Aug 2021 21:26:29 GMT'}, 'RetryAttempts': 0}}
The file spark_4_emr_codes_extraction.py contains the code for following paragraphs 4.3.1
The file spark_4_emr_I94_processing.py contains the code for following paragraphs 4.3.2, 4.3.3, 4.3.4
4.3.2 Coded fields: I94CIT and I94RES¶
I94CIT, I94RES contain codes indicating the country where the applicant is born (I94CIT), or resident (I94RES).
The data is extracted from I94_SAS_Labels_Descriptions.SAS. This can be done sporadically or every time a change occurred, for example a new code has been added.
The conceptual flow below was implemented.

First steps are define credential to access S3a then load the data in a dataframe, in a single row
AWS_ACCESS_KEY_ID = config['AWS']['AWS_ACCESS_KEY_ID']
AWS_SECRET_ACCESS_KEY = config['AWS']['AWS_SECRET_ACCESS_KEY']
hadoopConf = spark.sparkContext._jsc.hadoopConfiguration()
hadoopConf.set("fs.s3a.access.key", AWS_ACCESS_KEY_ID)
hadoopConf.set("fs.s3a.secret.key", AWS_SECRET_ACCESS_KEY)
text_file = 'I94_SAS_Labels_Descriptions.SAS'
text_file = os.path.join('s3a://', S3_BUCKET, S3_LAKE_RAWDATA, text_file)
df_label_wholetext = spark.read.text(text_file, wholetext=True)
Find the section of the file where I94CIT and I94RES are specified.
It start with I94CIT & I94RES and finish with the semicolon character.
To match the section, it is important to have the complete text in a single row, I did this using the option wholetext=True in the previous dataFrame read operation
@udf
def match_section(filetext, pattern):
# pattern = '(\/\* I94CIT & I94RES[^;]+)'
match = re.search(pattern, filetext)
return match.group(0)
pattern = "(\/\* I94CIT & I94RES[^;]+)"
df_I94CIT_RES = df_label_wholetext.withColumn('I94CIT&RES', match_section("value", lit(pattern))).select("I94CIT&RES")
Now I can split in a dataframe with multiple rows
df_I94CIT_RES_multi_rows = df_I94CIT_RES.withColumn('I94CIT&RES2', explode(split("I94CIT&RES", "[\r\n]+"))).select('I94CIT&RES2')
df_I94CIT_RES_multi_rows.show(5, truncate=False)
I filter the rows with structure \ = \
And then create 2 differents columns with code and country
@udf
def parse_country_name(line):
pattern = "(?<=')([0-9A-Za-z ,\-()]+)(?=')"
match = re.search(pattern, line)
if match:
return match.group(0)
else:
return ''
df_I94CIT_RES_multi_rows_filtered = df_I94CIT_RES_multi_rows.filter(df_I94CIT_RES_multi_rows['I94CIT&RES2'].rlike("([0-9]+ *\= *[0-9A-Za-z ',\-()]+)"))\
.withColumn('code', regexp_extract(df_I94CIT_RES_multi_rows['I94CIT&RES2'], "[0-9]+", 0))\
.withColumn('country', parse_country_name(col('I94CIT&RES2')))\
.drop("I94CIT&RES2")
df_I94CIT_RES_multi_rows_filtered.show(5,truncate=False)
I can finally store the data in a single file in json format
PROC_DATA_JSON = config['S3']['PROC_DATA_JSON']
file_pathname = 'country_codes'
# S3_bucket_country_codes = 'data/S3bucket_temp/country_codes' #for local testing
S3_bucket_country_codes = os.path.join('s3a://',S3_BUCKET, PROC_DATA_JSON, file_pathname )
# print(S3_bucket_country_codes)
df_I94CIT_RES_multi_rows_filtered.write.mode("overwrite").format('json').save(S3_bucket_country_codes)
4.3.3 Coded field: I94PORT¶
Similarly to extract the I94PORT codes
@udf
def match_section(filetext, pattern):
# pattern = '(\/\* I94CIT & I94RES[^;]+)'
match = re.search(pattern, filetext)
return match.group(0)
pattern = "(\/\* I94PORT[^;]+)"
df_I94PORT = df_label_wholetext.withColumn('I94PORT', match_section("value", lit(pattern))).select("I94PORT")
df_I94PORT_multi_rows = df_I94PORT.withColumn('I94PORT2', explode(split("I94PORT", "[\r\n]+"))).select('I94PORT2')
df_I94PORT_multi_rows_filtered = df_I94PORT_multi_rows.filter(df_I94PORT_multi_rows['I94PORT2'].rlike("([0-9A-Z.' ]+\t*\=\t*[0-9A-Za-z \',\-()\/\.#&]*)"))\
.withColumn('code', regexp_extract(df_I94PORT_multi_rows['I94PORT2'], "(?<=')[0-9A-Z. ]+(?=')", 0))\
.withColumn('city_state', regexp_extract(col('I94PORT2'), "(?<=\t')([0-9A-Za-z ,\-()\/\.#&]+)(?=')", 0))\
.withColumn('city', split(col('city_state'), ',').getItem(0))\
.withColumn('state', split(col('city_state'), ',').getItem(1))\
.withColumn('state', regexp_replace(col('state'), ' *$', ''))\
.drop('I94PORT2')
df_I94PORT_multi_rows_filtered.show(5,truncate=False) file_pathname = 'port_codes' # S3_bucket_port_codes = 'data/S3bucket_temp/port_codes'
S3_bucket_port_codes = os.path.join('s3a://',S3_BUCKET, PROC_DATA_JSON, file_pathname )
d4f_I94PORT_multi_rows_filtered.write.mode("overwrite").format('json').save(S3_bucket_port_codes)
The complete code for codes extraction is in spark_4_emr_codes_extraction.py
4.3.4 Data cleaning¶
The cleaning steps have already been shown in section 2, here are only summarized
- Load dataset
- Set schema
- Numeric fields: double to integer
- Fields dtadfile and dtaddto: string to date
- Fields arrdate and depdate: sas to date
- Handle nulls: no fill is set by default
- Drop duplicate
test_env = True
I94_TEST_FILE = '../../data/18-83510-I94-Data-2016/i94_apr16_sub.sas7bdat'
# load single file, entire dataset
if test_env:
df_I94 = spark.read.format('com.github.saurfang.sas.spark').load(I94_TEST_FILE).persist()
else:
df_I94 = spark.read.format('com.github.saurfang.sas.spark').load('{}/*/*.*'.format(I94_DATASET_PATH)).persist()
# modifying schema: Double => Integer
toInt = udf(lambda x: int(x) if x!=None else x, IntegerType())
df_I94 = df_I94.select( [ toInt(colname).alias(colname) if coltype == 'double' else colname for colname, coltype in df_I94.dtypes])
# modifying schema: string to date
df_I94 = df_I94.withColumn('dtadfile', to_date(col('dtadfile'), format='yyyyMMdd'))\
.withColumn('dtadddto', to_date(col('dtaddto'), format='MMddyyyy'))
@udf(TimestampType())
def to_timestamp_udf(x):
try:
return pd.to_timedelta(x, unit='D') + pd.Timestamp('1960-1-1')
except:
return pd.Timestamp('1900-1-1')
# modifying schema: sas to date
df_I94 = df_I94.withColumn('arrdate', to_date(to_timestamp_udf(col('arrdate'))))\
.withColumn('depdate', to_date(to_timestamp_udf(col('depdate'))))
# handle null, duplicate
fill_value = False
df_I94 = df_I94.na.fill(fill_value) #null
df_I94 = df_I94.drop_duplicates() #duplicates
4.3.5 Save clean data (parquet/json) to S3¶
%%time
is_local = False
PROC_DATA = config['S3']['PROC_DATA']
I94_DATA = config['S3']['I94_DATA']
if is_local:
S3_bucket_I94 = 'data/S3bucket_temp/I94_data' #for local testing
else:
S3_bucket_I94 = os.path.join('s3a://', S3_BUCKET, PROC_DATA, I94_DATA)
df_I94.write.format('parquet').mode('overwrite').partitionBy('i94yr', 'i94mon').save(S3_bucket_I94)
The complete code, refactorized and modularized, is in **spark_4_emr_I94_processing.py**
As a side note, saving the test file as parquet takes about 3 minute on the provisioned cluster. The complete script execution takes 6 minutes.
4.3.6 Loading, cleaning and saving airport codes¶
#load the data from the shared location
df_airport_codes = spark.read.csv('data/csv/airport-codes_csv.csv', header=True, inferSchema=True)
#dropping duplicates
df_airport_codes = df_airport_codes.drop_duplicates()
#selecting the save path
AIRPORT_CODES = config['S3']['AIRPORT_CODES']
if is_local:
S3_airport_codes = "data/S3bucket_temp/airport_codes"
else:
S3_bucket_I94 = os.path.join('s3a://',S3_BUCKET, PROC_DATA, AIRPORT_CODES)
#save parquet
df_airport_codes.write.parquet(S3_airport_codes, mode="overwrite")
4.4 Querying data on-the-fly¶
The data in the data lake can be queried on-place. That is the Spark cluster on EMR is directly operating on S3 data.
There are two possible ways to query the data:
- using Spark dataframe functions
- using SQL on tables
We see example of both programming styles.
These are some typical queries that are run on the data:
- For each port, in a given period, how many arrivals there are in each day?
- Where are the I94 applicants coming from, in a given period?
- In the given period, what port registered the highest number of arrivals?
- Number of arrivals in a given city for a given period
- Travelers genders
- Is there a city where the difference between male and female travelers is higher?
- Find most visited city (the function)
The queries are collected in the Jupyter notebook Capstone project 1 – Querying the data lake.ipynb
4.5 Querying data using the SQL querying style¶
df_I94.createOrReplaceTempView('DF_I94_TABLE')
spark.sql("""
SELECT COUNT(*)
FROM DF_I94_TABLE
""").show()
4.6 Data Quality Checks¶
The query-in-place concept implemented here uses a very short pipeline, data are loaded from S3 and after a cleaning process are saved as parquet. Quality of the data is guaranteed by design.
5. Write Up¶
The project has been set up with scalability in mind. All components used, S3 and EMR, offer higher degree of scalability, either horizontal and vertical.
The tool used for the processing, Apache Spark, is the de facto tool for big data processing.
To achieve such a level of scalability we sacrified processing speed. A data warehouse solution with a Redshift database or an OLAP cube would have been faster answering the queries. Anyway nothing forbids to add a DWH to stage the data in case of a more intensive, real-time responsive, usage of the data.
An important part of an ELT/ETL process is automation. Although it has not been touched here, I believe the code developed here is prone to be automatized with a reasonable small effort. A tool like Apache Airflow can be used for the purpose.
Scenario extension¶
- The data was increased by 100x.
In an increased data scenario, the EMR hardware needs to be scaled up accordingly. This is done by simply changing configuration in the config.cfg file. Apache Spark is the tool for big data processing, and is already used as the project analityc tool.
- The data populates a dashboard that must be updated on a daily basis by 7am every day.
In this case an orchestration tool like Apache Airflow is required. A DAG that trigger Phython scripts and Spark jobs executions, needs to be scheduled for daily execution at 7am.
The results of the queries for the dashboard can be saved in a file.
- The database needed to be accessed by 100+ people.
A proper database wasn’t used, on the contrary Amazon S3 is used to store data and queries them in-place. S3 is designed to massive scale in mind, it is able to handle sudden traffic spikes. Therefore, accessing the data by many people shouldn’t be an issue.
The programming used in the project, provision an EMR cluster for any user that plan to run it’s queries. 100+ EMRs is probably going to be expensive for the company. A more efficient sharing of processing resources must be realized.
6. Lessons learned¶
emr 5.28.1 use Python 2 as default¶
- As a consequence important Python packages like pandas are not installed by default for Python 3.
- To install them add an action to the EMR bootstrapping that install all packets
- install packages for Python 3: python 3 -m pip install \
Adding jars packages to Spark¶
For some reason adding the packages in the Python programm when instantiating the sparkSession doesn’t work (error message package not found).
This doesn’t work:
spark = SparkSession.builder\
.config("spark.jars","https://repo1.maven.org/maven2/com/epam/parso/2.0.8/parso-2.0.8.jar,https://repos.spark-packages.org/saurfang/spark-sas7bdat/2.0.0-s_2.11/spark-sas7bdat-2.0.0-s_2.11.jar")\
.config("spark.jars.packages","org.apache.hadoop:hadoop-aws:2.7.0")\
.enableHiveSupport()\
.getOrCreate()The packages must be added in the spark-submit:
spark-submit -jars https://repo1.maven.org/maven2/com/epam/parso/2.0.8/parso-2.0.8.jar,https://repos.spark-packages.org/saurfang/spark-sas7bdat/2.0.0-s_2.11/spark-sas7bdat-2.0.0-s_2.11.jar s3://helptheplanet/code/spark_4_emr_I94_processing.pyDebugging Spark on EMR¶
While evrything work locally, it doesn’t necessarily means that is going to work on the EMR cluster. Debugging the code is easier with SSH on EMR.
Reading an S3 file from Python is tricky¶
While reading with Spark is straightforward, one just needs to give the address s3://…., with Python boto3 must be used.
Transfering file to S3¶
During the debbuging phase, when the code on S3 must be changed many time using the web interface is slow and unpractical (permanently delete ). Memorize this command:aws s3 cp <local file> <s3 folder>
Removing the content of a directory from Python¶
import shutil
dirPath = 'metastore_db'
shutil.rmtree(dirPath)
7. References¶
- AWS CLI Command Reference
- EMR provisioning is based on: Github repo Boto-3 provisioning
- Boto3 Command Reference
